2. Haide Institute of Tropical Agricultural Resources (HITAR), Sanya, 572025, China
3. Alkali Soil Natural Environmental Science Center (ASNESC), Northeast Forestry Uinversity, Harbin, 150040, China
* These authors contributed equally to this work
 Author
Author  Correspondence author
Correspondence author
Bioscience Methods, 2010, Vol. 1, No. 2 doi: 10.5376/bm.2010.01.0002
Received: 01 Oct., 2010 Accepted: 24 Nov., 2010 Published: 27 Dec., 2010
Liu et al., 2010, Expression and Purification of Bacillus thuringiensis Cry1Ac22 protein in Escherichia coli, Bioscience Methods, 2010, Vol. 1, No. 2
A cry1Ac22 gene was amplified by PCR from Bacillus thuringiensis strain W015-1 isolated from diapausing larvae of silkworm (Bombyx mori). The full-length gene was ligated into the prokaryotic expression vector pQE30 to construct the recombinant plasmid pQE30-Cry1Ac22. The pQE30-Cry1Ac22 was transformed into competent cell of E.coli host strain M15 and then induced by IPTG to express His-tag-Cry1Ac22 fusion protein. The results showed that the His-tag-Cry1Ac22 was highly heterologous expressed in the presence of inclusion bodies in E.coli cell. Inducing experiments with different IPTG concentrations and temperatures revealed that the optimum condition for the expression of the fusion protein was 1 mmol/L IPTG and 28℃. SDS-PAGE analysis demonstrated that the host with pQE30-Cry1Ac22 generated a 133 kD His-tag-Cry1Ac22 fusion protein. The His-tag-Cry1Ac22 fusion protein was purified with affinity chromatography on a Ni2+-NTA resin column. Larvacidal assays were performed and showed that the engineered bacterial lysate and purified protein exhibited high insecticidal activity against second instar larvae of Plutella xylostella. This study might provide a basis for the preparation of antibody and for the determination of insecticidal activity using heterologous Cry1Ac22 protein.
Bacillus thuringiensis can generate insecticidal crystal protein during sporulation. Since Schnep and Whiteley isolated the first Bt insecticidal crystal protein in 1981 (Schnepf et al., 1981), nearly five hundred cry genes have been cloned in the world, with the majority belonging to the Cry1 family of genes. The Cry1A genes are listed in nine subfamilies including cry1Aaã€cry1Abã€cry1Acã€cry1Adã€cry1Aeã€cry1Afã€cry1Agã€cry1Ahã€and cry1Ai. These contain a total 93 hylotypes, while there are 34 genes in the cry1Ac subfamily (Crickmore et al., 2011, http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/ Bt/).
Cry1Ac insecticidal crystal protein was first found in Bacillus thuringiensis spp Kurstaki strain HD-73, which produces bipyramidal crystal proteins during sporulation with molecular weights between 129 kD to 138 kD. Bt HD-73 is recognized as an important standard strain due to well documented researches. The extensive application of transgenic plants with the Cry1Ac gene for lepidopteran control has lead to acquired resistance in Lepidoptera to the Cry1Ac crystal protein and has attracted global attention. Therefore, it becomes a more urgent task to look for novel Bt strains and diverse insecticidal crystal proteins to avoid the development resistance.
Bt W015-1 strains were isolated from the guts of diapausing silkworm (Bombyx mori) by Haide Institute of Tropical Agricultural Resources (HITAR), and its insecticidal activity was greater than that of known HD-73 strains (Xie et al., 2010).
Bt W015-1 strains can synthesize crystalline inclusions during sporulation with a molecular mass of 133 kD. The crystal protein possesses different restriction enzymatic digesting sites compared to that of Bt strain HD-73.
In this research we constructed the prokaryotic expression vector, pQE30-Cry1Ac22, to heterologously express in Eescherichia coli M15 and purified the inclusion protein His-tag-Cry1Ac22 in order to understand the characteristics and functions of the CryAc22 insecticidal crystal protein.
1 Results and analysis
1.1 The construction and identification of pQE30-Cry1Ac22 prokaryotic expression vector
We amplified the Cry1Ac22 gene, in the length of about 3 500 bp, from the plasmid DNA of Bt strain W015-1 ligated to the sequencing vector named as pMD18-T-Cry1Ac22 which was identified by the restriction enzymes BamHâ… and Sal â… (Figure 1A).
Sequencing analysis confirmed the amplified gene to be nearly identical to the Cry1Ac22 deposited in the GenBank. We ligated the gene from the pMD18-T-Cry1Ac22 into the prokaryotic expression plasmid pQE30 to construct E.coli expression vector by cutting with the restriction enzyme BamHâ… and SaIâ… . (Figure 1B). Further, the recombinant plasmid was validated by the restriction enzyme BamHâ… and named pQE30-Cry1Ac22 (Figure 1B).
The targeted gene cry1Ac22 in the recombinant has a length of about 3.5 kb, while its vector pQE30 has a length of 3.4 kb. It is too similar in length to be distinguished by digestion with BamHâ… and SaIâ… followed by agarose electrophoresis. Instead, we used a single restriction enzyme, BamHâ… , to cut the recombinant to obtain a single band about 6.9 kb in length.
|
|
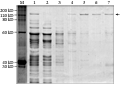
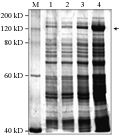
The pQE30-Cry1Ac22 plasmids were transformed into competent cells of E.coli M15 and single positive colonies were identified by PCR (Figure 2). Figure 2 shows that lane 1 to lane 5 were positive transformants which confirmed that the recombinant plasmid had been transformed into host bacterial cells of E.coli. The transformed Escherichia coli harboring plasmid pQE30-Cry1Ac22 was induced with 1 mmol/L IPTG to produce the recombinant proteins and 7.5â„… polyacrylamide gel electrophoresis was performed on the induced lysate to identify the inclusion protein. The results of SDS-PAGE showed that a distinct band of about 133 kD in molecular weight existed in the induced bacteria lygate transformed with pQE30-Cry1Ac22, whereas the band was lacking in the control without induction (Figure 3). The results indicated that recombinant plasmid pQE30-Cry1Ac22 should produce fusion protein 6×His-Cry1Ac22 under the induction of IPTG, and that the expressed amount of the fusion protein was increased gradually with the increase of inducing time (Figure 3).
|
|
|
|
In order to stabilize harvest of the expressed fusion proteins, we optimized the conditions for expressing the fusion proteins under temperature at 21℃, 28℃ and 37℃ and concentrations of IPTG at 0.1 mmol/L, 0.5 mmol/L and 1.0 mmol/L. The optimum condition was identified as 1.0 mmol/L IPTG concentration at 28℃ based on the results of SDS-PAGE (Figure 4).
|
|
1.2 Expression of fussion protein in E. coli M15
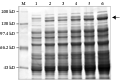
We conducted an experiment for the expression of Cry1Ac22 inclusion protein under conditions of 120 r/min shaking culture at 28℃ and induced by IPTG 1.0 mmol/L for 3 h, 6 h, 9 h, 12 h to observe the difference in protein expression among the trans- formants (Figure 5). The results showed that the amount of the expressed proteins was gradually increased with the length of ITPG induction. The different transformants exhibited their different expression levels under the fixed culture condition induced for 12 h (Figure 6).
|
|
|
|
1.3 Purification of the fusion protein
We purified the inclusion proteins expressed in E.coli using the urea-denaturation procedures. Bacterial lysate (before purification) and fractionated peak eluent (after purification) which both contained target proteins were collected and analyzed by SDS PAGE. The target protein band presented in the range of about 133 kD indicating that the fusion protein would be the dominant component in the profile of whole proteins in E.coli. The purity of the protein fractionated with Ni2+-NTA resin is about 80% (Figure 7). The concentration of the protein was detected to be 1 177 μg/mL.
|
|
1.4 Larvacide assay
Engineered bacterial lysate cultured for 20 hours and purified fusion protein were diluted and sprayed on Chinese cabbage leaves. Second instar larvae of diamondback moth, Plutella xylostella were introduced onto the leaves and held in an incubator at 25℃. The results showed that all larvae on treated leaves were dead after 30 hours, whereas the control larvae, normally, meaning that the engineered bacteria and its purified protein are toxic to the lepidopteran (data not shown).
2 Discussion
Making a prokaryotic fusion gene construct is a common and effective way to express an exogenous gene in E.coli. The prokaryotic expression vector pQE30 we used in this research is the most popular plasmid vector, having the characteristics of small molecular size (3.4 kb) and easy manipulation. It also has excellent features in its plasmid structures such as the replicon and ampicillin encoding gene from pBR322, the intense T5 promoter and the purifying tag that encodes six histidines (Qiagen, 2001).
E.coli M15 is the host strain that specificly matche with the pQE30 vector to express foreign protein. Using this strain can express exogenous gene to generate 6×His tagging inclusion protein. Fusion protein expressed in E.coli M15 usually has good stability and is not easily degraded by bacterial protease and the expressed inclusion proteins are easily fractionated by Ni2+-NTA resin affinity chromatography (Huang et al., 2008).
Inclusion body is the major form of foreign protein expressed in the bacteria, whose protein agglutinates in the host cell to develop active granules in sizes of 0.5~1 micron (μ). Inclusion body is insoluble in water while easy dissolved in some denaturants such as urea and hydrochloric acid (Haacke et al., 2008). The formation of inclusion badies will facilitate the high expression of foreign protein and prevent protein degradation by proteinase as well as avoiding poisoning the host cell from foreign protein (Hao et al., 1996).
In the present research, Cry1Ac22 fusion protein expressed in E.coli exists in the form of inclusion bodies. These are a kind of toxic protein existing in the host cell which reduces the concentration of the foreign protein and alleviates poisoning the host cells, increases the amount of protein expression. We think that purifying Cry1Ac22 protein using urea as denaturant will little affect the structure and function, which will be helpful to further study the physic- chemical characteristics and in vitro toxicity.
3 Materials and Methods
3.1 Strains, Plasmids and chemicalls
Details of the strains and plasmids used in this experiment are listed in Table 1. Taq DNA polyrmerase, dNTP, DNA ladder marker and λDNA/Hindâ…¢ marker were bought from DEMei Biotechnology Co. Ltd; Salâ… and BamHâ… were from TaKaRa Firm. Ni2+-NTAn resin is the production of Novagen Company.
|
|
3.2 Cry1Ac22 PCR amplification
A pair of primers was designed for amplifying Cry1Ac22 gene based on the sequence deposited in GenBank with accession no Eu282379, while BamHâ… and Salâ… restriction enzymic cutting sites were introduced into the termination of the primers. The forward primer is as GGA TCC ATG GAT AAC AAT CCG AAC ATC, and the reverse primer as GTC GAC TGA GTT TGC ATG AGA CTA TTC. The underlined letters refer to the restriction enzymic cutting sites of BamHâ… and Salâ… . The target gene was PCR amplified using plasmid DNA from Bt W015-1 under the conditions of PCR amplification as follows: Pre-denaturation at 95℃ for 5 min, followed by 30 cycles (94℃ 30 s, 54℃ 30 s and 72℃ 30 s), and finished at 72℃ for 5 min.
3.3 Construction procedures of recombinant expression vector
The enzymic cutting vector pQE30 and amplified Cry1Ac22 were mixed at a ratio of 3 moles to 1 mole in the ligation reaction mixture and deionized water was added to a final volume 20 μL. Prior to transformation, the mixture was held overnight in an ice bath at 16℃ to make pQE30-Cry1Ac22. 4 μL ligation product were placed in an ice bath with defrosting M15 competent cells for 30 min, prior to heat shock at 42℃ for 90 s, and then 400 μL LB medium were added and the culture was shaken for 1 hour at 37℃. 100 μL of the transformed liquid was placed on LB screening plates with 100 μg/mL ampicillin and 50 μg/mL kanamycin at 37℃ for overnight culture. Single colonies were picked and used for PCR to identification of positive recombinant clones.
3.4 Expression of fusion protein His-tag-Cry1Ac22 induced and optimized
A positive single bacterial colony was incubated in 5 mL LB liquid culture medium which was shaken overnight at 37℃, and then diluted with fresh medium at a ratio of 1:50 to increase the culture. Recombinant protein was then induced by adding 1.0 mmol/L IPTG at 37℃ when the OD600 reached about 0.8. 1 mL samples of culture medium were collected every 30 min.
The induced strains were collected by centrifugation at 4 000 r/min for 20 min and, after discarding the supernatant, the pellet was frozen at -80℃ for 2 hours, before completely suspending by first adding 5 mL suspension liquid and continuing to add suspension liquid to a final volume of 15 mL. Lysozyme was then added at 1 mg/mL to digest while shaking for 30 min in the ice bath.
Conditions for optimun expression were carried out under the IPTG concentrations of 0.1 mmol/L, 0.5 mmol/L, 1.0 mmol/L and temperatures at 20℃, 28℃, 37℃. Lysates were collected by centrifugation at 10 000 r/min at 4℃ for 40 min and 1 mL supernatant used for preparing samples for SDS-PAGE detection (Bradford, 1976).
3.5 The purification of His-tag-Cry1Ac22 fusion protein
Affinity chromatography with Ni2+-NTA Resin Column was used to purify the His-tag-Cry1Ac22 fusion protein. Supernatants were collected by super speed centrifugation and the pellet was transferred onto the Ni2+-NTA Resin Column for 2 h. Collect Recombinant bacterium cells were collected and cooled in an ice bath for 15 min and then Buffer B was added to re-suspend the collected pellet. The suspension was stirred for at least 15 min, (maxmum 60 min) at room temperature until the lytic reaction was finished. Supernatants were collected by super speed centrifugation at 10 000 g for 20 to 30 min, then transferred onto the Ni2+-NTA Resin Column. Buffer C was initially used to wash the column while collecting the fractionation liquid for SDS-PAGE analysis. This was followed by four washings with Buffer D and a final four washings with Buffer E to collect the respective liquid fractions for analysis by SDS-PAGE.
3.6 Bioassary
Second instar larvae of Plutella xylostella were provided by HITAR (Haide Institute of Tropical Agricultural Resources). Larvacidal assays followed the procedures of Xie et al (2010).
Authors’ contributions
ZML and SKL are the persons who executed this research and prepared the manuscript experiment; YZL partly managed the project and was involved in data analysis. XJF is the principal investigator for this project in charge of conducting experimental design, data analysis and preparing the manuscript. All authors have read and consent to the final version of this paper.
Acknowledgements
Our sincere thanks to all the people who have provided their strong technological support and useful advice during our experiments including Dr X. Zhang from ASNESC, and Mrs W. Zhang and L. Xie from HITAR. We greatly appreciate Dr. Phil Grau, Sr. Entomologist of SynTech Research, for reading and revising the manuscript. Many thanks to two anonymous reviewers for their strict criticism on this paper. This work was supported by the China National Bt Collection Initiative project. In this paper, mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by authors or institutes or the University involved in this study.
References
Adang M.J., Staver M.J., Rocheleau T.A., Leighton J., Barker R.F., and Thompson D.V., 1985, Characterized full-length and truncated plasmid clones of the crystal protein of Bacillus thuringiensis subsp. kurstaki HD-73 and their toxicity to Manduca sexta, Gene, 36(3): 289-300 doi:10.1016/0378-1119(85)90184-2
Bradford M.M., 1976, A rapid and sensitive method for the quantitation of microgram quantities utilizing the principle of protein dye binding, Anal. Biochem., 72(6): 248-254 doi:10.1016/0003-2697(76)90527-3
Crickmore et al., 2011, http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/
Haacke A., Fendrich G., Ramage P., and Geiser M., 2008, Chaperone over-expression in Escherichia coli: Apparent increased yields of soluble recombinant protein kinases are due mainly to soluble aggregates, Protein Expr. Purif., 64(2): 185-193 PMid:19038347
Hao H., Li H., 1996, The expression and purification of heterogeneous proteins synthesized in Escherichia coli, Chinese Journal of Biochemical Pharmaceutics, 17(5): 223-225
Huang S.F., Liu D.B., Zeng J.M., Xiao Q., Luo M., Zhang W.P., Tao K., Wen J.P., Huang Z.G., Feng W.L., 2008, Cloning, expression, purification and functional characterization of the oligomerization domain of Bcr-Abl oncoprotein fused to the cytoplasmic transduction peptide, Protein Expr. Purif., 64(2):167-178 PMid:19041400
Qiagen, ed.,2001, The QIAexpressionist. A handbook for high-level expression and purification of 6xHis-tagged proteins, Hilden, Germany, pp.1-128
Sambrook J.E., Fritsch F., Maaiatis T., 2002, Experiment guidance of molecular clone, Science press, Beijing, China, pp.483-485
Schnepf H.E., and Whiteley H.R., 1981, Cloning and expression of the Bacillus thuringiensis crystal protein gene in Escherichia coli, Proc. Nati. Acad. Sci., USA, 78(5): 2893-2897 doi:10.1073/pnas.78.5.2893
Xie L., Zhang W.F., Liu Z.M., Cai Y.G., Li Y.Z., Fang X.J., 2010, Characterization of a new highly toxic isolate of Bacillus thuringiensis from the diapausing larvae of silkworm and identification of cry1A 22 gene, Bt Research, Vol 2010 No 1 (DOI: 10.5376/bt.2010.01.0001)
. PDF(633KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Zhuoming Liu
. Shenkui Liu
. Youzhi Li
. Xuanjun Fang
Related articles
. Bacillus thuringiensis W015-1
. Cry1Ac22
. Fusion protein
. Heterologous expression
. in vitro Purification
. Larvacidal assay
Tools
. Email to a friend
. Post a comment
.png)